Infoboss
Bio-Information System
Conducting an integrated and systematic scientific analyses of Bio BigData
via collecting scattered heterogeneous Bio BigData and adopting a data model for Bio BigData
with novel AI technologies.
SOLUTION

About IBIS
Conducting an integrated and systematic
scientific analyses of Bio BigData via collecting
scattered heterogeneous Bio BigData and
adopting a data model for Bio BigData with
novel AI technologies.
Bio BigData
Analyses + AI technologies
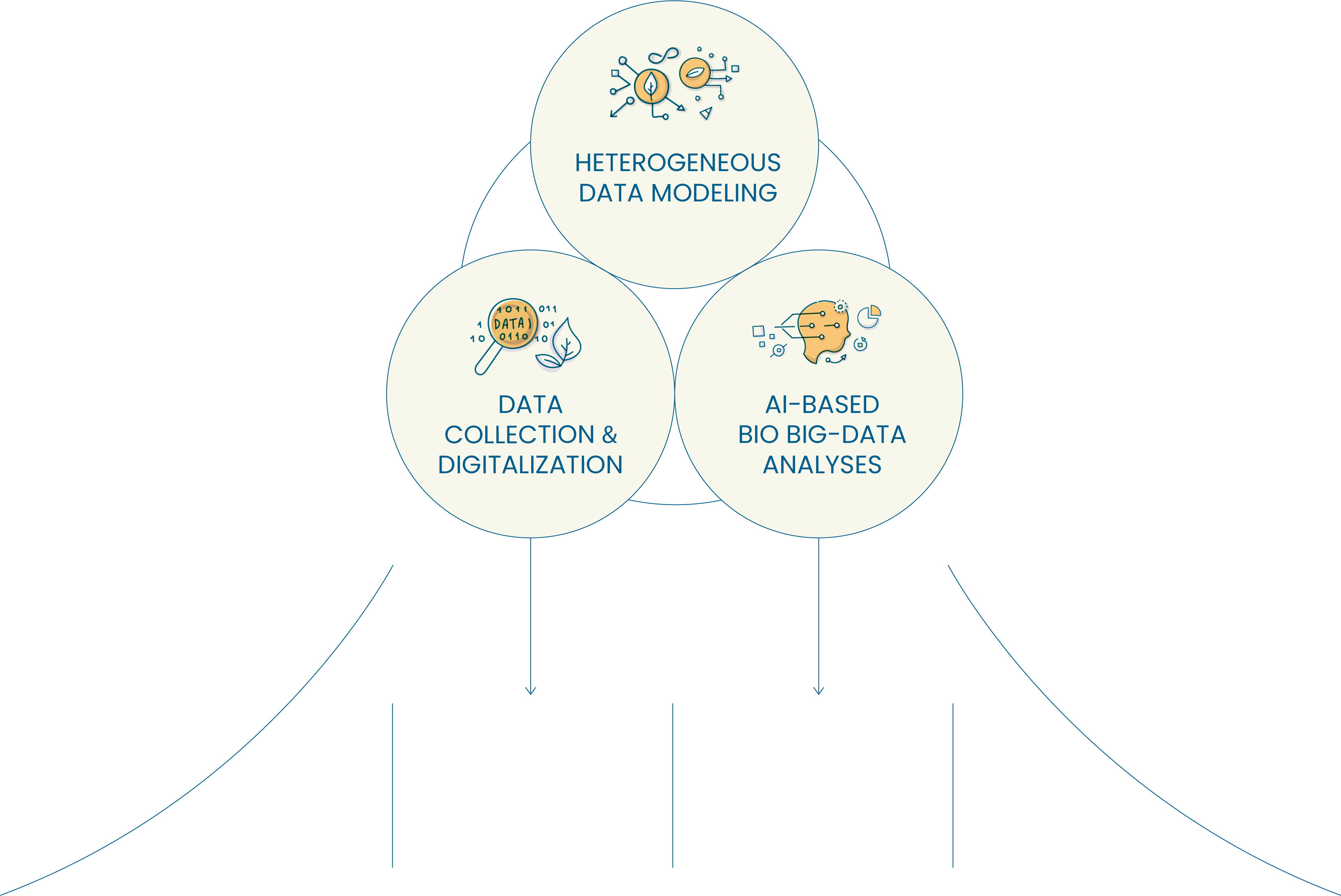
Data Model
Discovery and utilization
of novel information via
integration of heterogenous
Bio BigData
for Bio BigData
Development of
novel AI algorithms for
analyzing Bio BigData
including genome sequences
Meta-Series


INFOBOSS SOLUTIONS : Meta-Series
AI-based Meta-Series is optimized for
discovering novel native plant resources and
for their utilization via predicting useful
compounds from whole genomes.

-
This system collects and standardizes scattered and heterogeneous data including research papers, patents, and books with the standard species database, named as ISDB™. Furthermore, artificial intelligence (AI) algorithms were applied for analyzing these heterogeneous Bio BigData.
-
 UIBR®
UIBR®
The Useful Information for Biological Resources (UIBR®) aims to discover useful biological resources by integrating and standardizing information on their various uses. UIBR® collects useful information for biological resources scattered in books, old literatures, reports, and published papers. It integrates terminologies, such as local names, scientific names, drug names of biological resources, and disease names, with the standardized species list provided by ISDB™.
-
 UIBR-AI®
UIBR-AI®
AI platform for Useful Information for Biological Resources (UIBR-AI®) is the platform for predicting novel usages of plant resources with the aid of machine learning and deep learning algorithms based on the massive and diverse data stored in the UIBR® as well as related databases, including ISDB™.
-
 ISDB™
ISDB™
ISDB™ is an integrated system for managing species information originated from diverse species databases containing scientific names and synonyms. Collected data were integrated into the standardized species list as a standardized species data structure.
-
 RefDB™
RefDB™
Infoboss Reference Database (RefDB™) is an integrated database for managing and analyzing publications originated from various publication databases, such as PubMed. It contains i) various crawlers to collect references periodically, ii) the pipeline to build non-redundant reference information as well as iii) the web-based environment to choose and to analyze proper publications interested. In addition, RefDB™ provides the standardized reference information for many databases and systems of Infoboss.
-
 PAP™
PAP™
Patent Analysis Platform (PAP™) is an integrated platform for patents. Several crawlers for collecting patents efficiently have been implemented in this system. Additional analysis of patents was conducted together with RefDB™.
-
 MetaPool™
MetaPool™
MetaPool™ is the platform for classifying publications and patents originated from RefDB™ and PAP™, respectively, with the aids of machine learning and deep learning algorithms.
-
 MetaEve-AI™
MetaEve-AI™
MetaEve-AI™ was designed for evaluating economical values of plant resources in various aspects with the aid of the machine learning engine. It will provide the guide of many unused plant resources which have potential to be commercialized.

-
-
-
 GeIS™ (Genome Information System)
GeIS™ (Genome Information System)
Genome Information System (GeIS™) is the optimized system containing the four main pipelines for analyzing whole genome sequences efficiently. Various analysis tools implemented in the four pipelines can be executed through the GeIS™ web interface and the analysis results can be accessed and reanalyzed via the GlobalScrap®.
-
 GenomeArchive®
GenomeArchive®
GenomeArchive® is a standardized genome database which covering whole genome and organelle genome sequences. GenomeArchive® covers all organism's genome sequences from virus to human. GenomeArchive® also supports the multiple versions of genome sequences by providing previous version of sequences. All genetic elements, such as gene, transcript, and proteins, were accessible via genome browser. Modular design allows GenomeArchive® to provide these databases and interfaces to various system for supporting establishment of systems utilizing genome sequences cost effectively. Currently, 14,198 whole genomes and 3,180 organelle genomes were collected in GenomeArchive®.
-
 SSRDB™
SSRDB™
SSRDB™ was designed for identifying and analyzing simple sequence repeats from the whole and organelle genome sequences managed by GenomeArchive®. It contains the identification pipeline and various web-based analysis tools of SSRs. In addition, modular design of SSRDB™ provides the way to present identified SSRs in the different systems efficiently.
-
 Gene Family Database™
Gene Family Database™
Gene Family Database™ is a collection of independent gene family databases, such as GATA-TFDB, P450DB, and OSC-DB. Gene Family Database provides a standardized pipeline protocol to identify specific gene family with various filtering conditions and analysis pipelines including sequence alignment and construction of phylogenetic tree. In addition, basic format of web interfaces will provide the easy access to identified gene family members. Moreover, GlobalScrap® will provide the versatile environment to analyze them in detail to understand their characteristics. Two publications (R1 and R2) have been published based on this pipeline till now.

-
-
The phytocompounds predicted by this system will be verified in the Experimental Validation System (ExVS) for commercialization.
-
 IPDB™
IPDB™
Infoboss Pathway Database (IPDB™) was constructed for managing scattered biochemical reaction data with the non-redundant compound names and enzyme classifications. The pipeline named as Reaction Maker constructs the global reaction maps as well as compound-specific pathways. These generated pathways are utilized for predicting secondary metabolites from the whole genome sequences together with MetaPre-AI®. Additional biochemical pathways are also integrated into the IPDB™ for uncovering biochemical pathways of all phytocompounds.
-
 PhytoCompDB™
PhytoCompDB™
The Phytocompound Database (PhytoCompDB™) aims for standardizing phytocompounds with proper species, parts, and conditions with precise references. It can provide global view of phytocompounds based on the accumulated researches so far. We will try to utilize these identified phytocompounds in various aspects, including combining pathway information as well as whole genome data. We will also try to integrate the databases which has alrady established with specific purposes for expanding the coverage of phytocompound data.
-
 MetaPre-AI®
MetaPre-AI®
MetaPre-AI® is the system to predict phytocompounds from the plant whole genome sequences with ensemble engine containing traditional identification pipeline as well as multiple pipelines developed based on machine learning and deep learning.
-
 PECT™
PECT™
Platform for EC Number Prediction Tools (PECT™) is an integrated platform for analyzing enzyme sequences. Currently, 31,120,928 enzyme sequences were collected from several major database including BRENDA. Several classifiers for predicting enzyme functions as EC number were implemented to evaluate their accuracy for genes predicted from newly sequenced genome sequences. Moreover, annotation pipeline of EC number was also implemented to annotate and to feed as a training dataset of the classifier. Web interface and GlobalScrap® provide the analysis environment on the web.
-
 MetaTarget®
MetaTarget®
MetaTarget® was designed for predicting mode of actions as well as their biological activities of phytocompounds. In the case of prediction of mode of actions, our engine was constructed with consideration of mixtures, which is plant extract. For training these engines, published papers, molecular mechanisms including signal transduction pathway, related gene sequences, phytocompounds, as well as protein-small molecule interactions were utilized.

-
-
-
 Meta-ISM®
Meta-ISM®
Meta-ISM® was designed for confirming various phytocompounds from plant extracts with small number of HPLC-based experiments. It will be used for confirming phytocompounds which were predicted from the whole genome sequences by MetaPre-AI®.
-
 MetaEx™
MetaEx™
MetaEx™ was designed for confirming biological functions of plant extracts or phytocompounds especially predicted by MetaTarget®. It consists of various cell-based experiments, so called as in vitro experiments, and in vivo experiments. Standardized experiments for each function, such as anti-inflammatory, are used for providing the training dataset of MetaTarget® for precise prediction of its biological functionalities.

-
Meta-Doctor® is an advanced solution designed to accelerate complex clinical workflows through its robust AI-powered engine, seamlessly integrated with the Meta-Series solutions. By leveraging cutting-edge AI technologies, it optimizes the design of clinical trials and patient data analysis, enhances diagnostic accuracy, and streamlines administrative tasks. Additionally, Meta-Doctor® facilitates further valuable clinical processes based on data acquired from current clinical trial results. This not only significantly reduces the time and costs associated with clinical processes but also improves overall efficiency and patient outcomes.
Others
-
Biodiversity Information System Biodiversity is a research area to deal with heterogeneous Bio BigData. Infoboss Inc. provides the four specialized systems for managing and analyzing Bio BigData.
-
 Plant Management System (PMS™)
Plant Management System (PMS) is the system for providing management tools of plant resources especially in arboretums and gardens. PMS uses the standardized species list provided by ISDB™ for providing precise scientific names. Moreover, various functions for managing plant resources, such as import and export of plant species, were developed. All functions are available on the web site with multiple-user systems with several statistics functions.
Plant Management System (PMS™)
Plant Management System (PMS) is the system for providing management tools of plant resources especially in arboretums and gardens. PMS uses the standardized species list provided by ISDB™ for providing precise scientific names. Moreover, various functions for managing plant resources, such as import and export of plant species, were developed. All functions are available on the web site with multiple-user systems with several statistics functions.
-
 Biodiversity Observation Database (BODB™)
Biodiversity Observation Database (BODB™) is a web-based platform for managing diverse observation data acquired from digital machines. Currently, BODB™ contains 74,083 individuals having 144,741 pictures (696.69 GB) collected from cell phone camera as well as digital equipments. BODB™ provides i) the community platform for collecting biodiversity data from many people systematically, ii) academical research platform together with BODB™'s facilities (e.g., Flora Database of Mt. Suri; http://flora.mtsuri.net/), and iii) web-based analysis environment with 10 tools implemented in GlobalScrap®. In near future, BODB™ will be connected to versatile databases which contain weather information, genomic databases, and even image classifiers for utilizing collected images and metadata of biological resources.
Biodiversity Observation Database (BODB™)
Biodiversity Observation Database (BODB™) is a web-based platform for managing diverse observation data acquired from digital machines. Currently, BODB™ contains 74,083 individuals having 144,741 pictures (696.69 GB) collected from cell phone camera as well as digital equipments. BODB™ provides i) the community platform for collecting biodiversity data from many people systematically, ii) academical research platform together with BODB™'s facilities (e.g., Flora Database of Mt. Suri; http://flora.mtsuri.net/), and iii) web-based analysis environment with 10 tools implemented in GlobalScrap®. In near future, BODB™ will be connected to versatile databases which contain weather information, genomic databases, and even image classifiers for utilizing collected images and metadata of biological resources.
-
 Integrated Flora / Fauna Database (IFD™)
Integrated Flora and Fauna Database (IFD™) is a standardized database for archiving and analyzing investigation data of flora and fauna. Usually, research data of flora and fauna were not officially published or not available as an electronic form. It indicates that these data should be collected manually and format of data should be various. Moreover, scientific names or even common names used in these researches were different from each other due to changes of them. To overcome these problems, IFD™ provides standardized form of flora and fauna investigation and utilize the recent species list provided by ISDB™ with synonym data. Based on it, various analysis tools were implemented together with GlobalScrap® for further in-depth analyses.
Integrated Flora / Fauna Database (IFD™)
Integrated Flora and Fauna Database (IFD™) is a standardized database for archiving and analyzing investigation data of flora and fauna. Usually, research data of flora and fauna were not officially published or not available as an electronic form. It indicates that these data should be collected manually and format of data should be various. Moreover, scientific names or even common names used in these researches were different from each other due to changes of them. To overcome these problems, IFD™ provides standardized form of flora and fauna investigation and utilize the recent species list provided by ISDB™ with synonym data. Based on it, various analysis tools were implemented together with GlobalScrap® for further in-depth analyses.
-
 Herbarium Database (Infoboss Cyber Herbarium™)
Infoboss Cyber Herbarium™ is a herbarium for managing plant specimen with digitalized pipeline. Goal of Infoboss Cyber Herbarium™ is analyzing detailed phenotypes of plants from specimen for understanding plant classification with digitalized data. Even though coverage of plant species is not so large as similar as other herbariums, quality of digitalized data not only for pictures but also for measuring data will provide new aspect for plant taxonomy. In addition, based on experience of management of species database in Korea, plant taxonomy can be reanalyzed in different points of view.
Herbarium Database (Infoboss Cyber Herbarium™)
Infoboss Cyber Herbarium™ is a herbarium for managing plant specimen with digitalized pipeline. Goal of Infoboss Cyber Herbarium™ is analyzing detailed phenotypes of plants from specimen for understanding plant classification with digitalized data. Even though coverage of plant species is not so large as similar as other herbariums, quality of digitalized data not only for pictures but also for measuring data will provide new aspect for plant taxonomy. In addition, based on experience of management of species database in Korea, plant taxonomy can be reanalyzed in different points of view.
-
-
Environmental Sensors (IoT) InfoStation/InfoNode® are the integrated system for collecting microenvironmental data
surrounded by organisms in nature.-
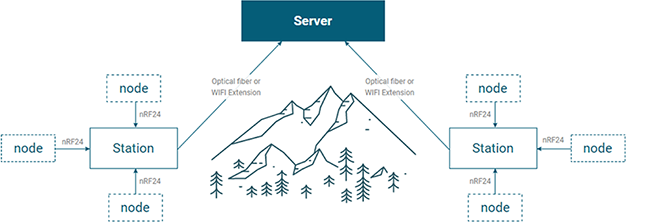
 InfoStation / InfoNode®
- InfoStation/InfoNode® are integrated environmental sensors specialized for natural habitats in mountain areas without electricity supply.
InfoStation / InfoNode®
- InfoStation/InfoNode® are integrated environmental sensors specialized for natural habitats in mountain areas without electricity supply.
- These are the integrated system for collecting microenvironmental data surrounded organisms in nature on the spot and in real-time.
- Collected microenvironmental data can be utilized for various industries after analyzing them.
-
-
IMS (Information Management System) Infoboss deals with various types of information including Bio Big-Data.
Information Management System (IMS) will provides the efficient ways to manage, analyze, and publish them in the standardized ways.-
 GlobalScrap®
- GlobalScrap® is the system for exchanging heterogeneous Bio BigData via web interfaces.
GlobalScrap®
- GlobalScrap® is the system for exchanging heterogeneous Bio BigData via web interfaces.
- It encapsulated heterogeneous data for free exchanges between systems.
- It also provides the common interface to execute any programs with the heterogeneous data which users are interested in. -
 Infoboss Gateway™
- Infoboss Gateway is the system to export data processed by systems in Infoboss Inc.
Infoboss Gateway™
- Infoboss Gateway is the system to export data processed by systems in Infoboss Inc.
- Various services, such as the utilization of bioinformatics tools managed by GeIS™ and published papers related to biological resources (RefDB™) have been provided through the Infoboss Gateway.
-



